Data#
Independently of the model, data will be needed for either creating or validating the model. The amount of data required will be determined by a trade-off between complexity-affordability-accuracy , and the type of model. Data-driven models will likely require a broad range and large volume of data, while mechanistic models may only need data for a defined set of variables.
1.1 Data Sources#
Berkeley Earth Global Temperature
Daily average temperature observations
Resolution 1° x 1°
Monthly mean temperature FAIRBANKS
80 km from Nenana, data from 1828 - 2020
Monthly mean temperature HEALY RIVER AIRPORT
90 km from Nenana, data from 1983 - 2012
Monthly mean temperature MOUNT MCKINLEY NATL
100 km from Nenana, data from 1923 - 2013
NOAA GHCN: Nenana
NOAA GHCN: Fairbanks
USGS Water Data: Nenana
USGS Water Data: Fairbanks
USGS Water Data: Fairbanks
NERC-EDS Global Cloud Coverage
TEMIS Global Solar Surface Irradiance
USGS Glaciers Data: Gulkana
NOAA Global Indexes
Nenana Ice Classic
Ice thickness measurements
Ice break up dates
1.2 Loading data#
If you are new to Python you are probably aware of some basic data structures like lists and dictionaries, maybe even NumPy arrays.
We will now introduce a new data structure, DataFrames.
DataFrames#
A DataFrame is tabular data structure that stores information in rows and columns, where each column has a name and can hold different types of data. Both the rows and columns can have labels, making it easier to access and manipulate specific part of the data (very colloquially, think of a dataframe as a mix between dictionaries and a spreadsheets)
In Python, we will use Pandas.Dataframes (quick introduction)
DataFrames are particularly useful because we can easily manipulate data using a wide variety built-in methods. Additionally, pandas.dataframe can easily handle data with timestamps, meaning, that we can assign a date and time to each observation and manipulate these variables without worrying about time zones, leap years, or other time-related issues. For example, you can subtract one date from another to find the number of days between them, access data for specific years or months, iterate through years, etc.
In addition to rows and columns, dataframe have an .index.The index is series of labels associated to each row, by default the index assign an integer, starting from 0, to each row.
When working with time-series it may be convenient to use a DatetimeIndex to assign a timestamp label to each row instead.
Task 1:
The files corresponding to the sources mentioned above are available in the repository in the folder data/raw_files, choose a file, load the contents to a Pandas.DataFrame, set the index to a datetime object and plot the timeseries.
Tips/Help
Read the documentation for pd.read_csv. Pay special attention to the following methods and arguments
pd.read_csv()
skip_row
sep
index_col
pd.index=pd.to_datetime()format
If you downloaded this notebook, you will need to download the file from the repository and pass the filepath to pd.read_csv.
If you cloned the whole repository, the data files can be easily loaded using the relative path ../../data/raw_files/file_name.txt
If you are running the notebook in your browser, use the function import_data_browser from the ice package to load the file into the browser. The function needs the URL of the file within the repository, specifically the raw URL. To obtain this, navigate to the file in the repository, and in the upper right-hand side, there is a button named Raw that will direct you to a page with the raw contents of the text file. The URL of this page is the URL that you need to pass to the function.
# EX 1.
# # Exercise 1.
import pandas as pd
import matplotlib.pyplot as plt
import iceclassic as ice
#Temp=pd.read_csv('../../data/raw_files/Berkeley temp.txt',skiprows=23,sep='\t',index_col=0)
file1=ice.import_data_browser('https://raw.githubusercontent.com/iceclassic/mude/main/book/data_files/Berkeley%20temp.txt')
Temp=pd.read_csv(file1,skiprows=23,sep='\t',index_col=0)
Temp.index=pd.to_datetime(Temp.index,format='%Y%m%d')
plt.figure(figsize=(20,5))
plt.plot(Temp)
plt.title("Temperature Data")
plt.xlabel("Date")
plt.ylabel("Temperature (C)")
plt.show()
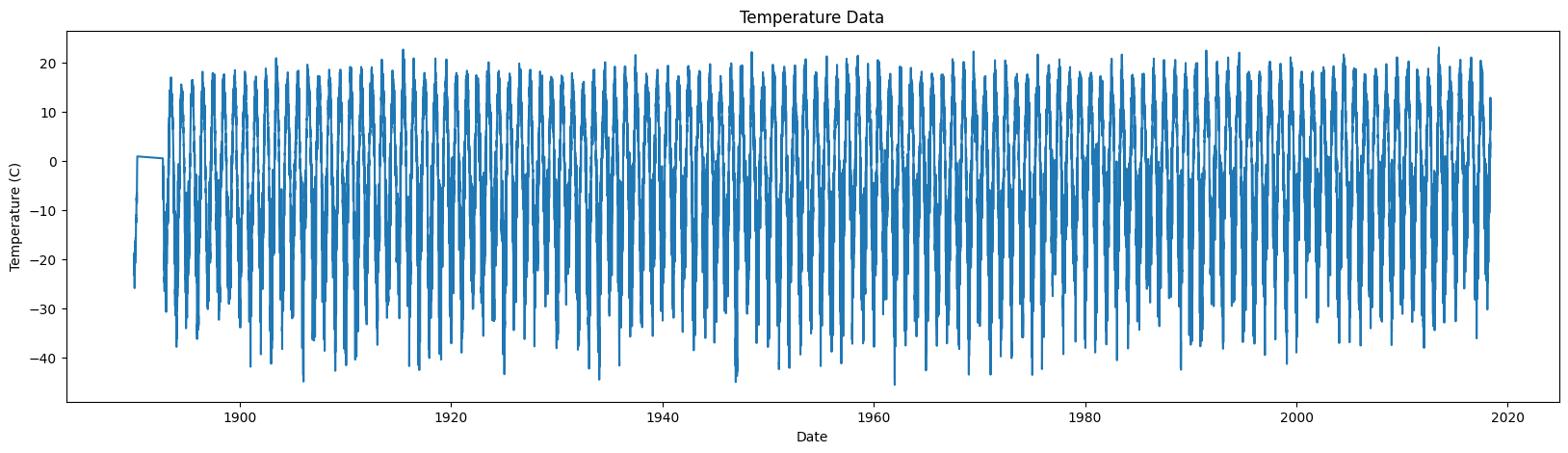
Task 2:
Choose another file, load it and merge it to the previously loaded file, make sure that the resulting DataFrame contains two columns and a datetime index.
Tips/Help
Read the documentation for pd.merge.
If we merge two dataframe (df1, df2) with datetime indexes, the observations (rows) kept in the resulting dataframe depend on the type merge, this is determine by the argument how.
- `how='inner'`: Only observation associted with dates present in both df1 and df2 are kept.
- `how='left'`: All observation from df1 are kept. Observation in df2 with dates that don’t match dates in df1 are filled with NA.
- `how='right'`: Similar to `how='left'`, but observations from second df are kept.
- `how='outer'`: All observation from both df1 and df2 are kept. Observation associated with non-matching dates are filled with NA.
# # Exercise 2.
# Loading 2nd file
#PDO=pd.read_csv('../../data/raw_files/PDO.csv',skiprows=1,index_col=0,sep=';')
file2=ice.import_data_browser('https://raw.githubusercontent.com/iceclassic/mude/main/book/data_files/PDO.csv')
PDO=pd.read_csv(file2,skiprows=1,index_col=0,sep=';')
PDO.index=pd.to_datetime(PDO.index,format='%Y%m')
# Merging
merged=pd.merge(Temp,PDO,left_index=True,right_index=True,how='outer')
merged.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 46500 entries, 1854-01-01 to 2020-02-01
Data columns (total 2 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 # TAVG [degree C] Air Surface Temperature 46018 non-null float64
1 Value 1994 non-null float64
dtypes: float64(2)
memory usage: 1.1 MB
1.3 Basic DataFrame manipulation#
The function pd.read_csv can loading most data files. However, dealing with files with varying use of separators, delimiters, data types and formats can be a time-consuming.
To simplify the process of loading and merging multiple DataFrames individually, all data sources have been merged into a single text file that can be easily loaded. The file can be found in data/Time_series_DATA.txt
The file has more than 20 variables (columns), some of which have daily observations spanning more than a century. To contextualize this variable the following interactive map shows the weather station where they were measured.
upload the files to github for this map, as the functions allows to choose the detail level we need a bunch of shapefiles
#ice.plot_interactive_map(plot_only_nearby_basin=False)
---------------------------------------------------------------------------
DataSourceError Traceback (most recent call last)
Cell In[5], line 1
----> 1 ice.plot_interactive_map(plot_only_nearby_basin=False)
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\iceclassic\example.py:1180, in plot_interactive_map(Pfafstetter_levels, plot_only_nearby_basin)
1178 # changing the level to higher number yield more basin, using Pfafstetter levels 1-12 source HydroBASINS
1179 file='../../data/shape_files/hybas_lake_ar_lev'+'{:02d}'.format(Pfafstetter_levels)+'_v1c.shp'
-> 1180 gdf_basin_lev = gpd.read_file(file)
1181 if plot_only_nearby_basin:
1182 if Pfafstetter_levels==1:
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\geopandas\io\file.py:294, in _read_file(filename, bbox, mask, columns, rows, engine, **kwargs)
291 from_bytes = True
293 if engine == "pyogrio":
--> 294 return _read_file_pyogrio(
295 filename, bbox=bbox, mask=mask, columns=columns, rows=rows, **kwargs
296 )
298 elif engine == "fiona":
299 if pd.api.types.is_file_like(filename):
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\geopandas\io\file.py:547, in _read_file_pyogrio(path_or_bytes, bbox, mask, rows, **kwargs)
538 warnings.warn(
539 "The 'include_fields' and 'ignore_fields' keywords are deprecated, and "
540 "will be removed in a future release. You can use the 'columns' keyword "
(...)
543 stacklevel=3,
544 )
545 kwargs["columns"] = kwargs.pop("include_fields")
--> 547 return pyogrio.read_dataframe(path_or_bytes, bbox=bbox, **kwargs)
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\pyogrio\geopandas.py:261, in read_dataframe(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, fid_as_index, use_arrow, on_invalid, arrow_to_pandas_kwargs, **kwargs)
256 if not use_arrow:
257 # For arrow, datetimes are read as is.
258 # For numpy IO, datetimes are read as string values to preserve timezone info
259 # as numpy does not directly support timezones.
260 kwargs["datetime_as_string"] = True
--> 261 result = read_func(
262 path_or_buffer,
263 layer=layer,
264 encoding=encoding,
265 columns=columns,
266 read_geometry=read_geometry,
267 force_2d=gdal_force_2d,
268 skip_features=skip_features,
269 max_features=max_features,
270 where=where,
271 bbox=bbox,
272 mask=mask,
273 fids=fids,
274 sql=sql,
275 sql_dialect=sql_dialect,
276 return_fids=fid_as_index,
277 **kwargs,
278 )
280 if use_arrow:
281 meta, table = result
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\pyogrio\raw.py:196, in read(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, return_fids, datetime_as_string, **kwargs)
56 """Read OGR data source into numpy arrays.
57
58 IMPORTANT: non-linear geometry types (e.g., MultiSurface) are converted
(...)
191
192 """
194 dataset_kwargs = _preprocess_options_key_value(kwargs) if kwargs else {}
--> 196 return ogr_read(
197 get_vsi_path_or_buffer(path_or_buffer),
198 layer=layer,
199 encoding=encoding,
200 columns=columns,
201 read_geometry=read_geometry,
202 force_2d=force_2d,
203 skip_features=skip_features,
204 max_features=max_features or 0,
205 where=where,
206 bbox=bbox,
207 mask=_mask_to_wkb(mask),
208 fids=fids,
209 sql=sql,
210 sql_dialect=sql_dialect,
211 return_fids=return_fids,
212 dataset_kwargs=dataset_kwargs,
213 datetime_as_string=datetime_as_string,
214 )
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\pyogrio\_io.pyx:1239, in pyogrio._io.ogr_read()
File c:\Users\gabri\anaconda3\envs\ice_package_test\Lib\site-packages\pyogrio\_io.pyx:219, in pyogrio._io.ogr_open()
DataSourceError: ../../data/shape_files/hybas_lake_ar_lev04_v1c.shp: No such file or directory
Task 3:
Read the documentation of explore_data() from the iceclassic package and use it to explore the contents of Time_series_DATA.txt.
Decide which variables might not be relevant to the problem ? Explain.
#Data=pd.read_csv("../../data/Time_series_DATA.txt",skiprows=149,index_col=0,sep='\t')
file3=ice.import_data_browser('https://raw.githubusercontent.com/iceclassic/mude/main/book/data_files/time_serie_data.txt')
Data=pd.read_csv(file3,skiprows=162,index_col=0,sep='\t')
Data.index = pd.to_datetime(Data.index, format="%Y-%m-%d")
ice.explore_contents(Data)
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 39309 entries, 1901-02-01 to 2024-02-06
Data columns (total 28 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Regional: Air temperature [C] 38563 non-null float64
1 Days since start of year 38563 non-null float64
2 Days until break up 38563 non-null float64
3 Nenana: Rainfall [mm] 29547 non-null float64
4 Nenana: Snowfall [mm] 19945 non-null float64
5 Nenana: Snow depth [mm] 15984 non-null float64
6 Nenana: Mean water temperature [C] 2418 non-null float64
7 Nenana: Mean Discharge [m3/s] 22562 non-null float64
8 Nenana: Air temperature [C] 31171 non-null float64
9 Fairbanks: Average wind speed [m/s] 9797 non-null float64
10 Fairbanks: Rainfall [mm] 29586 non-null float64
11 Fairbanks: Snowfall [mm] 29586 non-null float64
12 Fairbanks: Snow depth [mm] 29555 non-null float64
13 Fairbanks: Air Temperature [C] 29587 non-null float64
14 IceThickness [cm] 461 non-null float64
15 Regional: Solar Surface Irradiance [W/m2] 86 non-null float64
16 Regional: Cloud coverage [%] 1463 non-null float64
17 Global: ENSO-Southern oscillation index 876 non-null float64
18 Gulkana Temperature [C] 19146 non-null float64
19 Gulkana Precipitation [mm] 18546 non-null float64
20 Gulkana: Glacier-wide winter mass balance [m.w.e] 58 non-null float64
21 Gulkana: Glacier-wide summer mass balance [m.w.e] 58 non-null float64
22 Global: Pacific decadal oscillation index 1346 non-null float64
23 Global: Artic oscillation index 889 non-null float64
24 Nenana: Gage Height [m] 4666 non-null float64
25 IceThickness gradient [cm/day]: Forward 426 non-null float64
26 ceThickness gradient [cm/day]: Backward 426 non-null float64
27 ceThickness gradient [cm/day]: Central 391 non-null float64
dtypes: float64(28)
memory usage: 8.7 MB
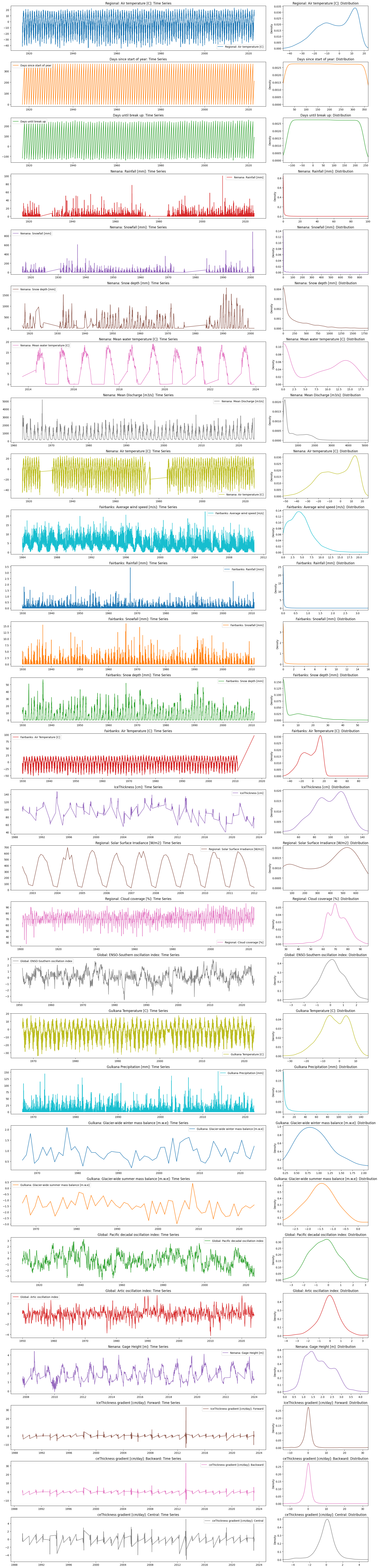
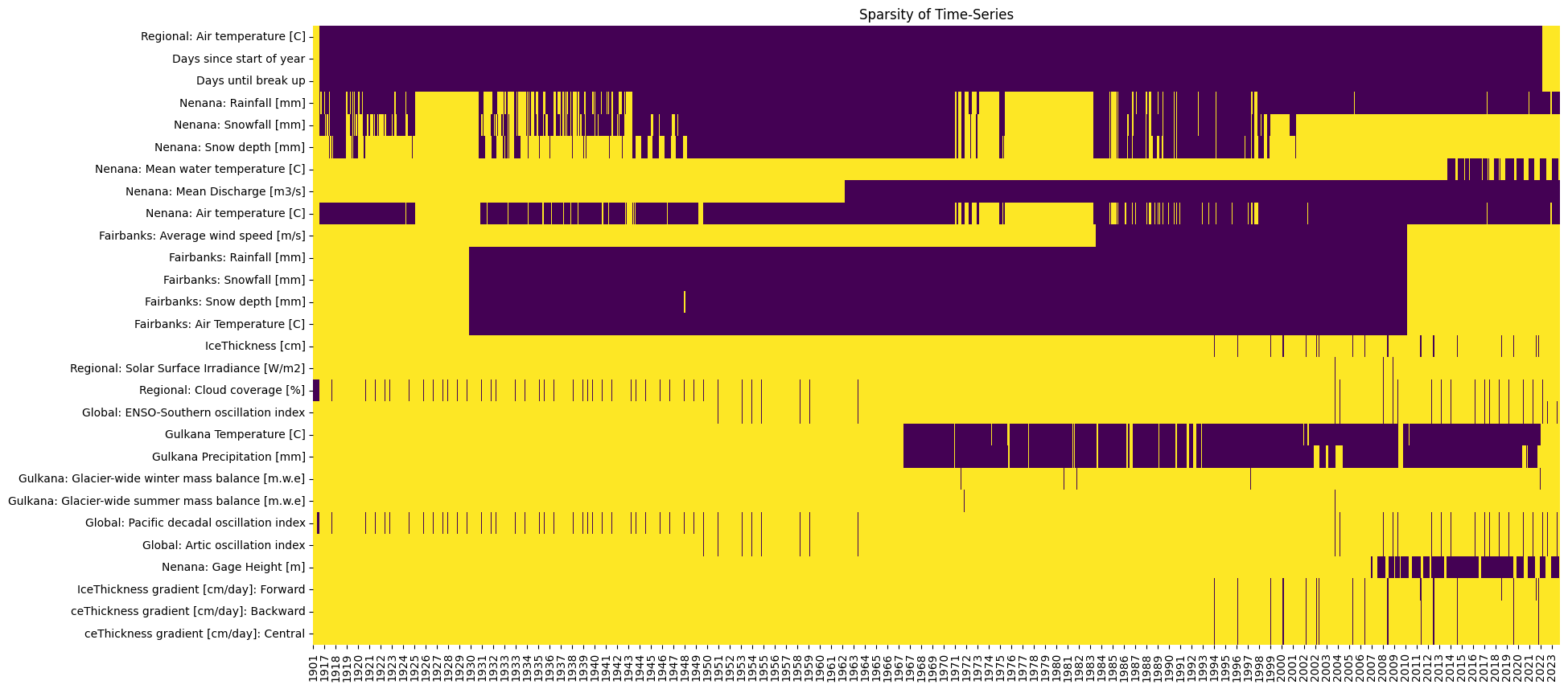
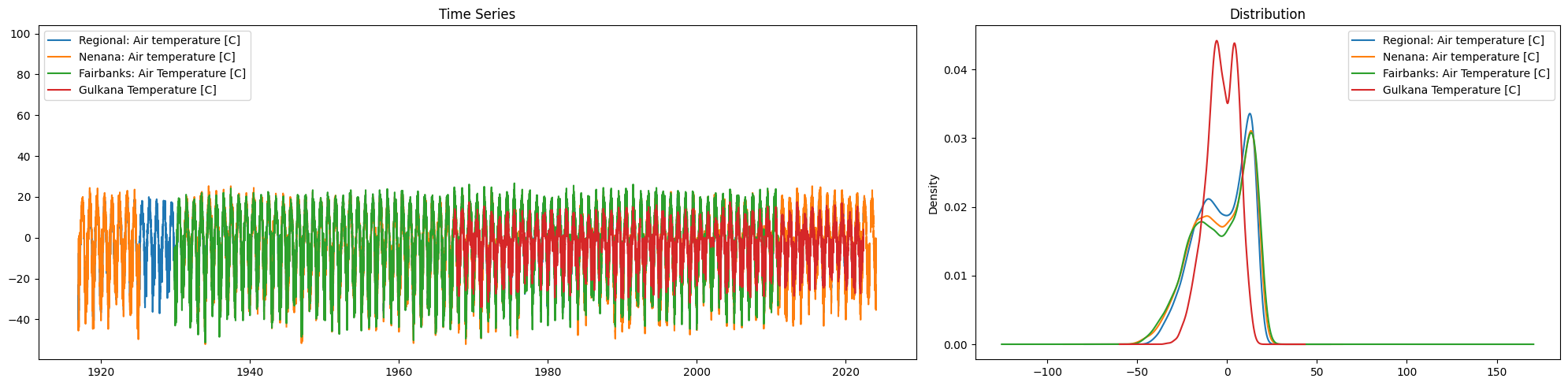
Task 4:
The file contains data for three distinct temperature timeseries, use compare_columns to visually compare them.
Then, use .drop() to eliminate the columns that might be redundant .
temperature_columns=['Regional: Air temperature [C]','Nenana: Air temperature [C]','Fairbanks: Air Temperature [C]','Gulkana Temperature [C]']
ice.compare_columns(Data,temperature_columns)
Data=Data.drop(columns=temperature_columns[1:3])
The contents of a DataFramecan be grouped into subsets of the dataframe by means of simple indexing.
There are three main ways to use indexing to create a subset of the dataframe
df.loc[]: uses the label of columns (column names) and rows (index) to select the subsetdf.iloc[]: uses integers indexes to select the subsetdf[]: uses the column name to select the column. Additionally we can pass a list/array of boolean values to use as a mask
Task 5:
Create a new dataframe with a subset containing only the data from 1950 onwards
Data_2 = Data[(Data.index.year >= 1950)]
Data_2.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 27065 entries, 1950-01-01 to 2024-02-06
Data columns (total 26 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Regional: Air temperature [C] 26510 non-null float64
1 Days since start of year 26510 non-null float64
2 Days until break up 26510 non-null float64
3 Nenana: Rainfall [mm] 22266 non-null float64
4 Nenana: Snowfall [mm] 13322 non-null float64
5 Nenana: Snow depth [mm] 12965 non-null float64
6 Nenana: Mean water temperature [C] 2418 non-null float64
7 Nenana: Mean Discharge [m3/s] 22562 non-null float64
8 Fairbanks: Average wind speed [m/s] 9797 non-null float64
9 Fairbanks: Rainfall [mm] 22250 non-null float64
10 Fairbanks: Snowfall [mm] 22250 non-null float64
11 Fairbanks: Snow depth [mm] 22250 non-null float64
12 IceThickness [cm] 461 non-null float64
13 Regional: Solar Surface Irradiance [W/m2] 86 non-null float64
14 Regional: Cloud coverage [%] 876 non-null float64
15 Global: ENSO-Southern oscillation index 876 non-null float64
16 Gulkana Temperature [C] 19146 non-null float64
17 Gulkana Precipitation [mm] 18546 non-null float64
18 Gulkana: Glacier-wide winter mass balance [m.w.e] 58 non-null float64
19 Gulkana: Glacier-wide summer mass balance [m.w.e] 58 non-null float64
20 Global: Pacific decadal oscillation index 889 non-null float64
21 Global: Artic oscillation index 889 non-null float64
22 Nenana: Gage Height [m] 4666 non-null float64
23 IceThickness gradient [cm/day]: Forward 426 non-null float64
24 ceThickness gradient [cm/day]: Backward 426 non-null float64
25 ceThickness gradient [cm/day]: Central 391 non-null float64
dtypes: float64(26)
memory usage: 5.6 MB
Task 6
Create a new DataFrame which contains only the columns ['Regional: Air temperature [C]','Nenana: Rainfall [mm]'], for the years [2008,2010], from jan-01 to may-01.
Filter the original DataFrame using masks.
Tips/Help
Use df.index.year to create a mask that filters the years and df.index.strftime() for the mask pertaining to the dates.
cols=['Regional: Air temperature [C]','Nenana: Rainfall [mm]']
years=[2008,2009,2010]
date_1='01/01'
date_2='03/21'
year_mask = Data.index.year.isin(years) # (df.index.year >= min(years)) & (df.index.year <= max(years)) is also an option
date_mask = (Data.index.strftime('%m/%d') >= date_1) & (Data.index.strftime('%m/%d') <= date_2)
filtered_df = Data.loc[year_mask & date_mask, cols]
plt.figure(figsize=(20,5))
plt.plot(filtered_df.index,filtered_df,alpha=0.4,marker='o')
plt.xlabel("Date")
plt.ylabel("Values")
plt.title("Filtered DataFrame")
plt.legend(filtered_df.columns)
plt.show()
1.4 Interactive Plot#
The DataFrame has decades of observations, which causes the xaxis(date) of the plot to lose detail, we could make the figure larger, or plot subset of the dataframe, alternatively we can use plot_columns_interactive() to create a plot were we can scroll and zoom at specific dates.
::{card} Ex 9
Read the documentation for plot_columns_interactive() from the iceclassic package and create an interactive plot.
change the location of the file to the -github path- the function internally uses a file wiht the break up dates, eaither change it or extract this date from the the big dataframe with
break_up_dates=Data.index[Data['Days until break up']==0]
column_groups = {
'Group 1': ['Regional: Air temperature [C]','Gulkana Temperature [C]'],
'Group 2': ['Nenana: Snow depth [mm]'],
'Group 3': ['Nenana: Mean Discharge [m3/s]']}
# Plot the specified columns with default y_domains and focus on a specific date
#ice.plot_columns_interactive(Data, column_groups, title="Break up times & Global Variables at Tenana River-Nenana, AK with break-up dates")
