MUDE W3- Gradient Estimation#
Info:
This notebook is a very simple draft showing how we could use data from the ~~Nenana Ice Classic~~ (from any of the column in our dateset) for the numerical modelling part of MUDE.
The implementation is not particularly efficient, but it is quite flexible.
The dataset used is direclty loaded from github, so a a few extra packages are needed(io and requests) but we could avoid using ‘non-standard’libraries by downloading the file and loading it ‘manually’.
from io import StringIO
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly.graph_objects as go
import plotly.express as px
import plotly
import iceclassic as ice
Loading the dataset#
# we could load the data from the repo
file=ice.import_data_browser('https://raw.githubusercontent.com/iceclassic/mude/main/book/data_files/time_serie_data.txt')
Data=pd.read_csv(file,skiprows=162,index_col=0,sep='\t')
Data.index = pd.to_datetime(Data.index, format="%Y-%m-%d")
#Data.info()
River_data=Data[['Nenana: Mean Discharge [m3/s]','Nenana: Gage Height [m]']]
#River_data.info()
River_data.dropna(inplace=True)
Ice_temp=Data[['IceThickness [cm]','Regional: Air temperature [C]']]
Ice_temp.dropna(how='all', inplace=True)
Ice_temp['Day']=Ice_temp.index.dayofyear
Ice_temp.info()
Ice_temp.to_csv('Ice&Temp.csv')
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 38578 entries, 1917-01-01 to 2023-04-21
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 IceThickness [cm] 461 non-null float64
1 Regional: Air temperature [C] 38563 non-null float64
2 Day 38578 non-null int32
dtypes: float64(2), int32(1)
memory usage: 1.0 MB
C:\Users\gabri\AppData\Local\Temp\ipykernel_13704\1496965268.py:8: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
River_data.dropna(inplace=True)
C:\Users\gabri\AppData\Local\Temp\ipykernel_13704\1496965268.py:12: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
Ice_temp.dropna(how='all', inplace=True)
C:\Users\gabri\AppData\Local\Temp\ipykernel_13704\1496965268.py:13: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
Ice_temp['Day']=Ice_temp.index.dayofyear
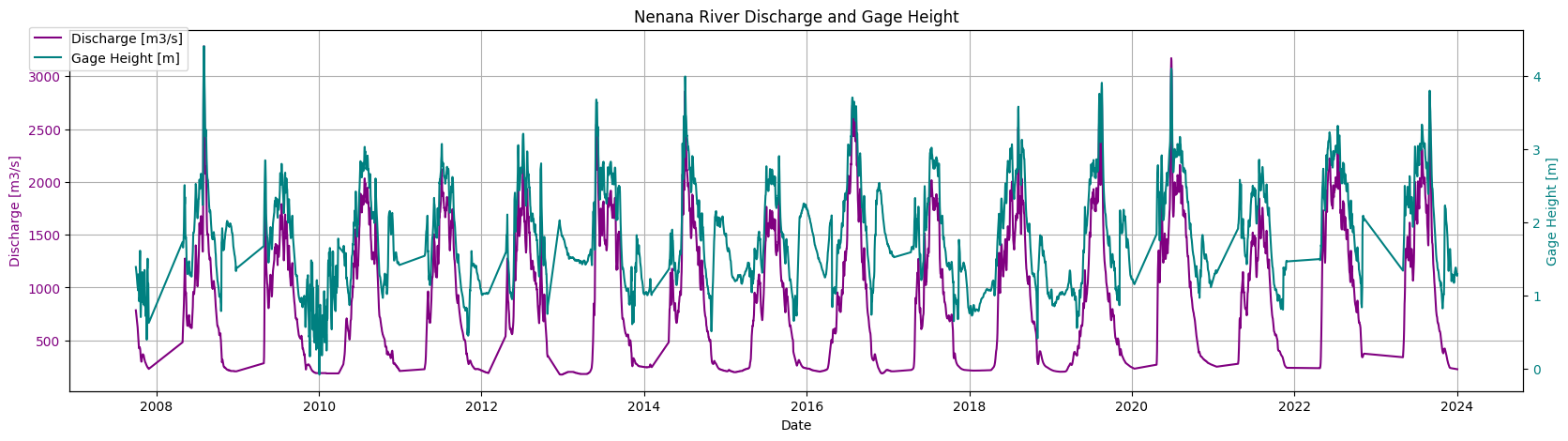
River_data=pd.read_csv('Nenana_River.csv',index_col=0)
River_data.index = pd.to_datetime(River_data.index, format="%Y-%m-%d")
fig, ax1 = plt.subplots(figsize=(20, 5))
ax1.set_title("Nenana River Discharge and Gage Height")
ax1.plot(River_data.index, River_data['Nenana: Mean Discharge [m3/s]'], color='purple', label='Discharge [m3/s]')
ax1.set_xlabel('Date')
ax1.set_ylabel('Discharge [m3/s]', color='purple')
ax1.tick_params(axis='y', labelcolor='purple')
ax1.grid()
# ax1.set_xlim(pd.Timestamp('2018-01-01'), pd.Timestamp('2018-12-31'))
ax2 = ax1.twinx()
ax2.plot(River_data.index, River_data['Nenana: Gage Height [m]'], color='teal', label='Gage Height [m]')
ax2.set_ylabel('Gage Height [m]', color='teal')
ax2.tick_params(axis='y', labelcolor='teal')
fig.legend(loc='upper left', bbox_to_anchor=(0.1, 0.9))
plt.show()
# we could load the data from the repo
file=ice.import_data_browser('https://raw.githubusercontent.com/iceclassic/mude/main/book/data_files/time_serie_data.txt')
Data=pd.read_csv(file,skiprows=162,index_col=0,sep='\t')
Data.index = pd.to_datetime(Data.index, format="%Y-%m-%d")
Data.info()
break_up_dates=Data.index[Data['Days until break up']==0]
print(str(break_up_dates[break_up_dates.year==2019]))
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 39309 entries, 1901-02-01 to 2024-02-06
Data columns (total 28 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Regional: Air temperature [C] 38563 non-null float64
1 Days since start of year 38563 non-null float64
2 Days until break up 38563 non-null float64
3 Nenana: Rainfall [mm] 29547 non-null float64
4 Nenana: Snowfall [mm] 19945 non-null float64
5 Nenana: Snow depth [mm] 15984 non-null float64
6 Nenana: Mean water temperature [C] 2418 non-null float64
7 Nenana: Mean Discharge [m3/s] 22562 non-null float64
8 Nenana: Air temperature [C] 31171 non-null float64
9 Fairbanks: Average wind speed [m/s] 9797 non-null float64
10 Fairbanks: Rainfall [mm] 29586 non-null float64
11 Fairbanks: Snowfall [mm] 29586 non-null float64
12 Fairbanks: Snow depth [mm] 29555 non-null float64
13 Fairbanks: Air Temperature [C] 29587 non-null float64
14 IceThickness [cm] 461 non-null float64
15 Regional: Solar Surface Irradiance [W/m2] 86 non-null float64
16 Regional: Cloud coverage [%] 1463 non-null float64
17 Global: ENSO-Southern oscillation index 876 non-null float64
18 Gulkana Temperature [C] 19146 non-null float64
19 Gulkana Precipitation [mm] 18546 non-null float64
20 Gulkana: Glacier-wide winter mass balance [m.w.e] 58 non-null float64
21 Gulkana: Glacier-wide summer mass balance [m.w.e] 58 non-null float64
22 Global: Pacific decadal oscillation index 1346 non-null float64
23 Global: Artic oscillation index 889 non-null float64
24 Nenana: Gage Height [m] 4666 non-null float64
25 IceThickness gradient [cm/day]: Forward 426 non-null float64
26 ceThickness gradient [cm/day]: Backward 426 non-null float64
27 ceThickness gradient [cm/day]: Central 391 non-null float64
dtypes: float64(28)
memory usage: 8.7 MB
DatetimeIndex(['2019-04-14'], dtype='datetime64[ns]', freq=None)
Estimating gradients#
the function below its included in the Pypi package, I’ll leave the code here for a while to double check the implementation is correct and I’ll remove it in a few days/week
def finite_differences(series:pd.Series): “”” Computes forward, central, and backward differences using the step size as days between measurement.
Parameters
---------
series: pd.Series
Series with datetime index
Returns
---------
df: pd.DataFrame
DataFrame with forward, backward and central differences for the Series
"""
days_forward = (series.index.to_series().shift(-1) - series.index.to_series()).dt.days
days_backward = (series.index.to_series() - series.index.to_series().shift(1)).dt.days
# Forward difference: (f(x+h) - f(x)) / h
forward = (series.shift(-1) - series) / days_forward
# Backward difference: (f(x) - f(x-h)) / h,
backward = (series - series.shift(1)) / days_backward
# Central difference: (f(x+h) - f(x-h)) / (h_forward + h_backward)
central = (series.shift(-1) - series.shift(1)) / (days_forward + days_backward) #
# fixing start/end points
forward.iloc[-1] = np.nan
backward.iloc[0] = np.nan
return pd.DataFrame({'forward': forward, 'backward': backward, 'central': central})
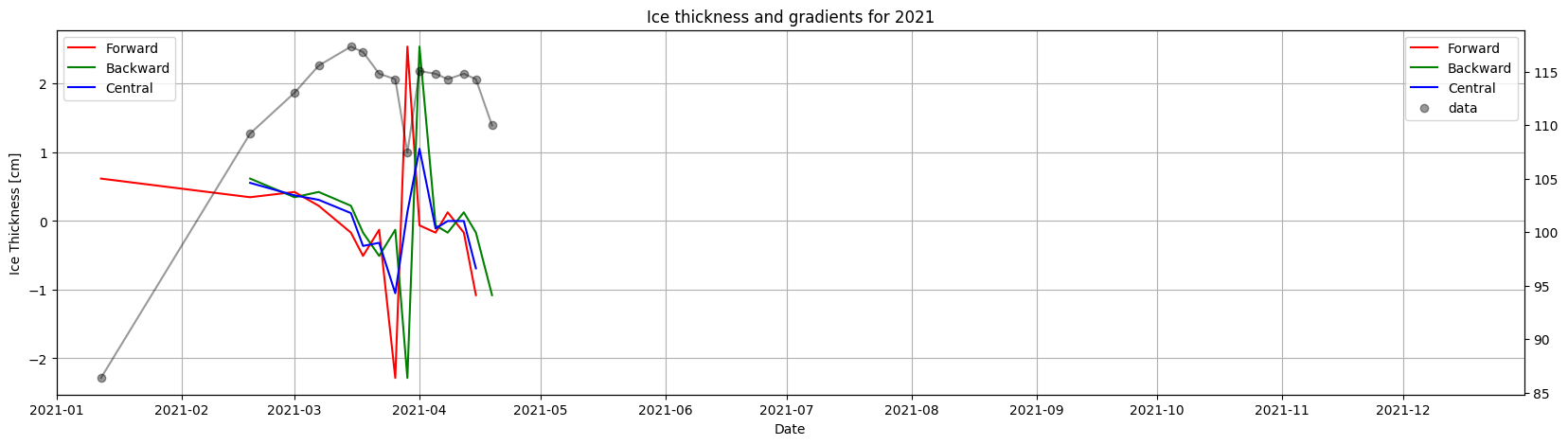
Ice thickness#
Ice_thickness=Data['IceThickness [cm]']
Ice_thickness=Ice_thickness.dropna(inplace=False)
print(Ice_thickness.head())
result = Ice_thickness.groupby(Ice_thickness.index.year,group_keys=True).apply(lambda x: ice.finite_differences(x)).reset_index(level=0, drop=True)
print(result)
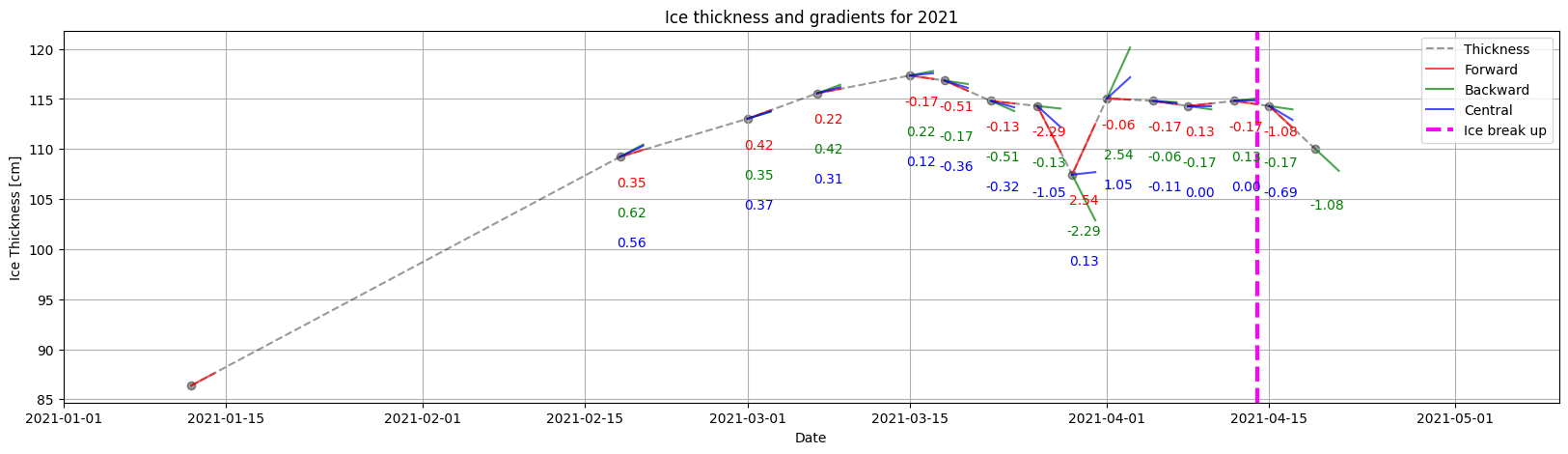
ice.plot_gradients_and_timeseries(result,
Ice_thickness,
2021,
Title='Ice thickness and gradients for 2021',
ylabel='Ice Thickness [cm]')
ice.plot_gradients_and_timeseries(result,
Ice_thickness,
year=2021,
plot_gradient_as_slope=True,
xlim=['01/01', '05/10'],
Title='Ice thickness and gradients for 2021',
ylabel='Ice Thickness [cm]',
vline={'Ice break up': break_up_dates[break_up_dates.year == 2019].strftime('%m/%d')[0]})
1989-02-26 106.68
1989-03-16 95.25
1989-03-21 95.25
1989-03-25 102.87
1989-03-28 105.41
Name: IceThickness [cm], dtype: float64
forward backward central
1989-02-26 -0.635000 NaN NaN
1989-03-16 0.000000 -0.635000 -0.496957
1989-03-21 1.905000 0.000000 0.846667
1989-03-25 0.846667 1.905000 1.451429
1989-03-28 0.181429 0.846667 0.381000
... ... ... ...
2023-04-07 1.439333 -1.270000 -0.108857
2023-04-10 -0.635000 1.439333 0.254000
2023-04-14 0.000000 -0.635000 -0.362857
2023-04-17 0.190500 0.000000 0.108857
2023-04-21 NaN 0.190500 NaN
[461 rows x 3 columns]
Discharge#
fix plotting function, so than when selecting an xlim, the function only compute the data in that range, right now it computes all the data and then restricts the plot which is slow
print(type(break_up_dates[break_up_dates.year == 2019].strftime('%m/%d')[0]))
Discharge=Data['Nenana: Mean Discharge [m3/s]']
Discharge=Discharge.dropna(inplace=False) # drop the missing values
result_2 = Discharge.groupby(Discharge.index.year,group_keys=True).apply(lambda x: ice.finite_differences(x))
result_2 = result_2.reset_index(level=0, drop=True)
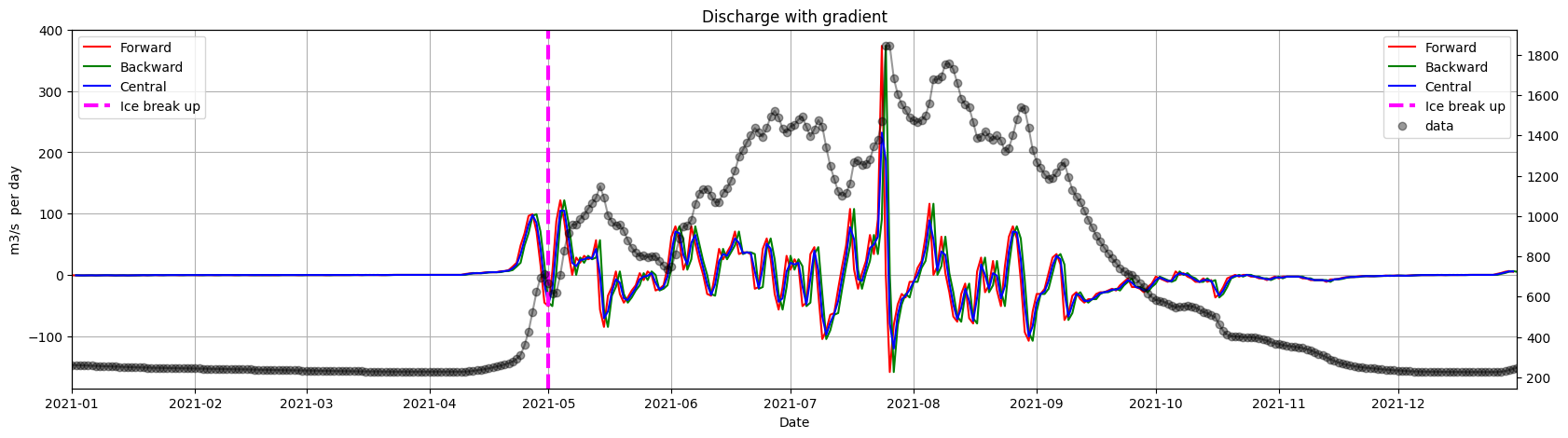
ice.plot_gradients_and_timeseries(result_2,
Discharge,
2021,
Title='Discharge with gradient',
ylabel='m3/s per day',
vline={'Ice break up': break_up_dates[break_up_dates.year == 2021].strftime('%m/%d')[0]})
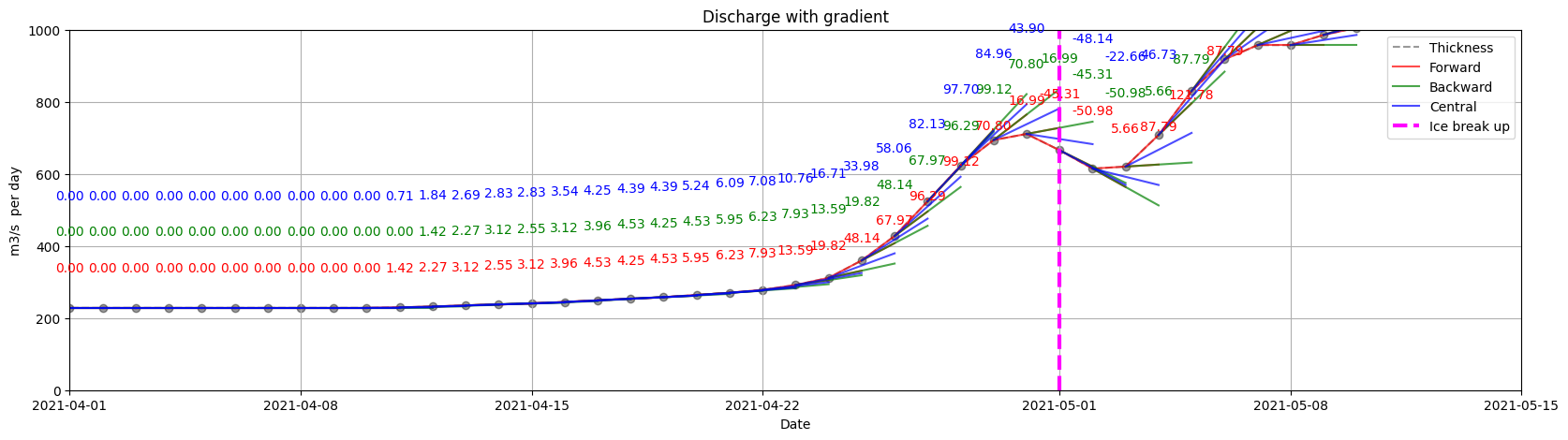
ice.plot_gradients_and_timeseries(result_2,
Discharge,
2021,
plot_gradient_as_slope=True,
xlim=['04/01', '05/15'],
Title='Discharge with gradient',
ylabel='m3/s per day',
ylim=[0, 1000],
annotation_offsets={'forward': 100, 'backward': 200, 'central': 300},
vline={'Ice break up': break_up_dates[break_up_dates.year == 2021].strftime('%m/%d')[0]})
#
<class 'str'>
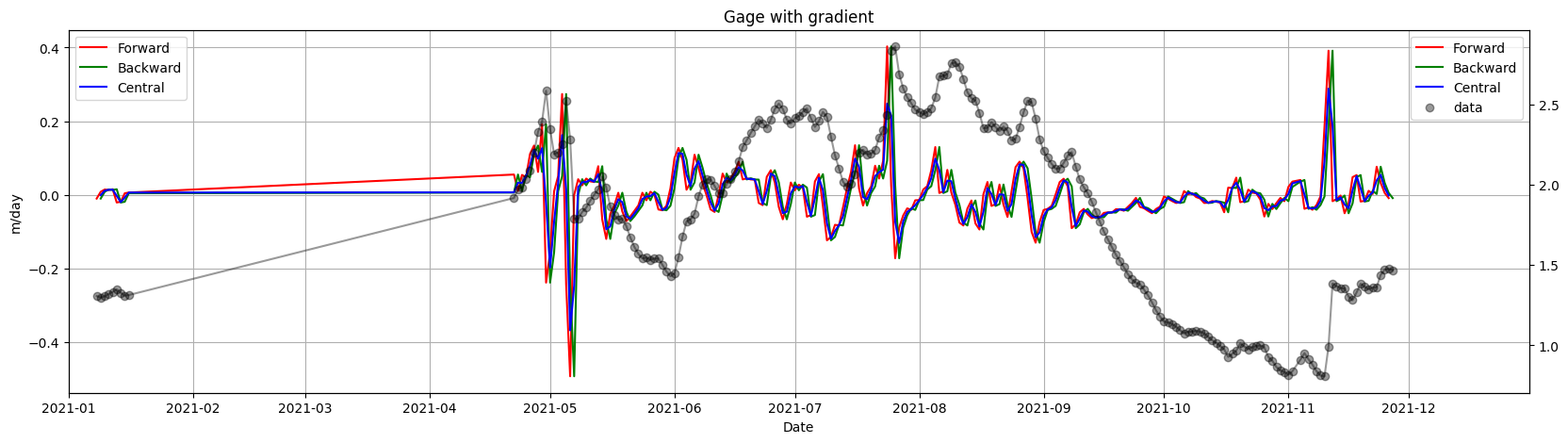
Gage Height#
Gage=Data['Nenana: Gage Height [m]']
Gage=Gage.dropna(inplace=False) # drop the missing values
result_3 = Gage.groupby(Gage.index.year,group_keys=True).apply(lambda x: ice.finite_differences(x))
result_3 = result_3.reset_index(level=0, drop=True)
ice.plot_gradients_and_timeseries(result_3,
Gage,
2021,
Title='Gage with gradient'
,ylabel='m/day')
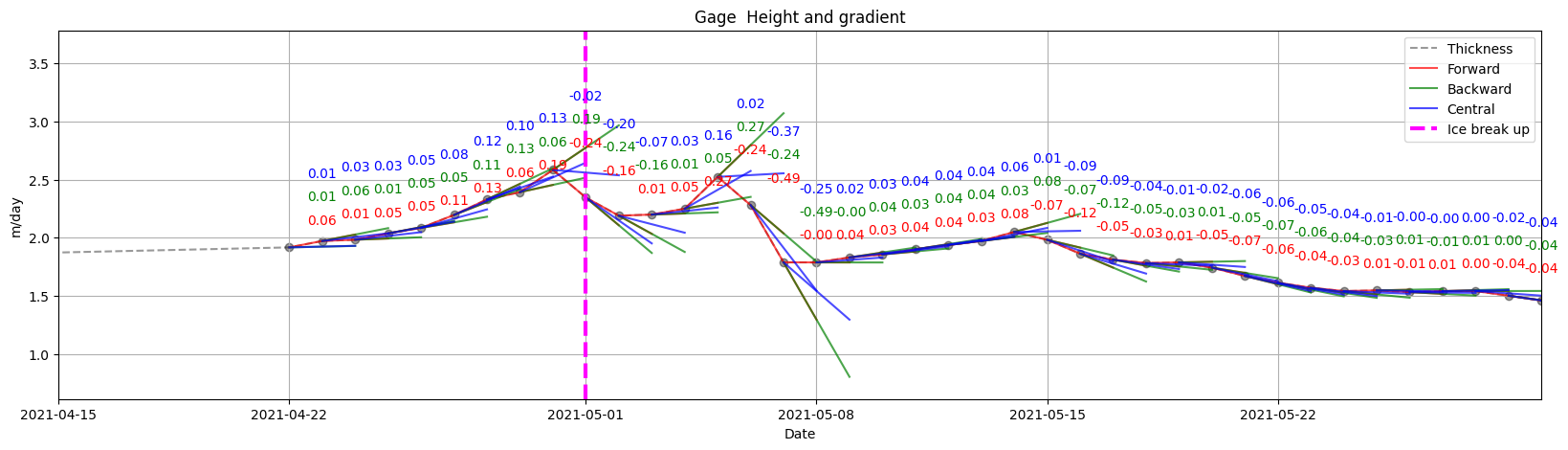
ice.plot_gradients_and_timeseries(result_3,
Gage,
2021,
plot_gradient_as_slope=True,
xlim=['04/15', '05/30'],
annotation_offsets={'forward': 0.2, 'backward': 0.4, 'central': 0.6},
ylabel='m/day',Title='Gage Height and gradient',
vline={'Ice break up': break_up_dates[break_up_dates.year == 2021].strftime('%m/%d')[0]})